Research
An organism’s genome contains all the information necessary for its development and function, including genes that encode RNA and proteins, regulatory regions that coordinate gene activity to form different cells and respond to stimuli, structural regions responsible for the stability, organization and faithful DNA transmission during cell division, as well as “parasitic” elements such as jumping genes (transposons) and regions of no or unknown role. It is essential that this information is organized and switched on and off in a controlled manner, so that only pieces relevant to a specific cell type and function are active at a given time, while irrelevant or harmful genetic elements are silenced. One of the most important means of genome regulation in eukaryotes is through the association of DNA with histone proteins into a higher order chromatin structure. 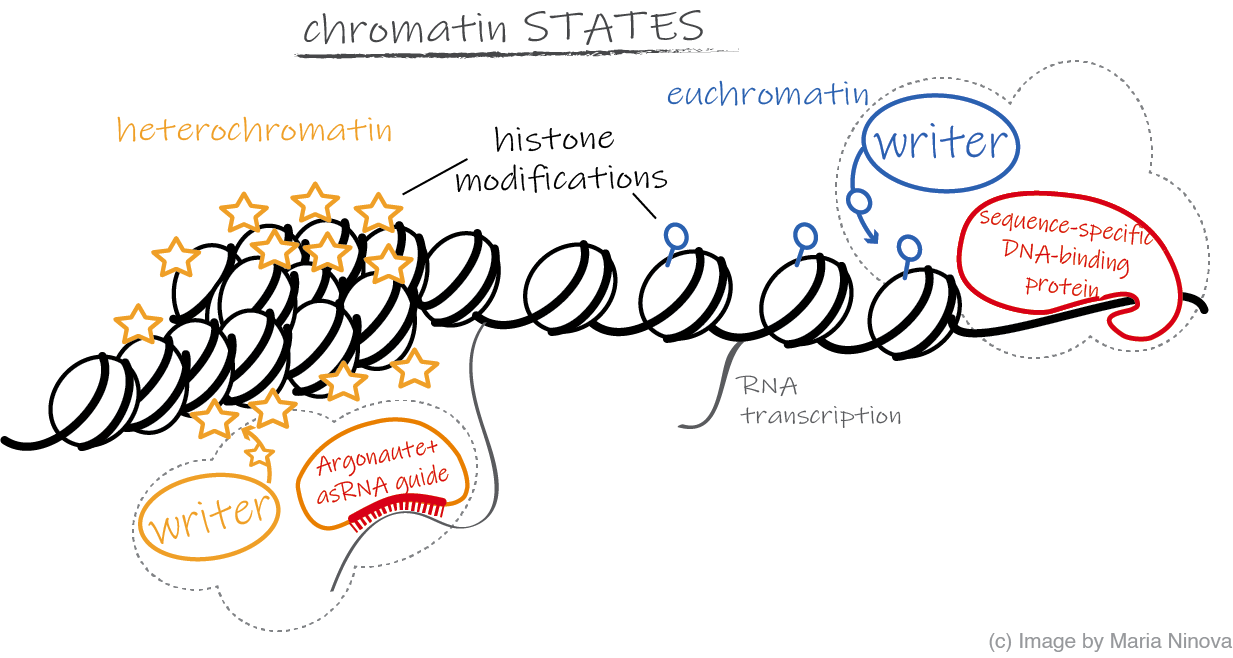 Post-translational histone modifications can alter chromatin density and recruit diverse downstream regulatory and structural factors that modulate gene activity. Our lab is interested in fundamental mechanisms that govern the formation of silencing chromatin marks at different genomic regions. We address these questions by a combiation of molecular biology, imaging, genomics (RNA-seq, ChIP-seq), and proteomics approaches. Most of our projects utilize the Drosophila melanogaster model system, which laid the foundations of modern genetics and has been at the frontier of chromatin, piRNA (see below), and transposon research. Areas of specific interest include (but are not limited to):
Post-translational histone modifications can alter chromatin density and recruit diverse downstream regulatory and structural factors that modulate gene activity. Our lab is interested in fundamental mechanisms that govern the formation of silencing chromatin marks at different genomic regions. We address these questions by a combiation of molecular biology, imaging, genomics (RNA-seq, ChIP-seq), and proteomics approaches. Most of our projects utilize the Drosophila melanogaster model system, which laid the foundations of modern genetics and has been at the frontier of chromatin, piRNA (see below), and transposon research. Areas of specific interest include (but are not limited to):
The mechanism of piRNA-mediated silencing
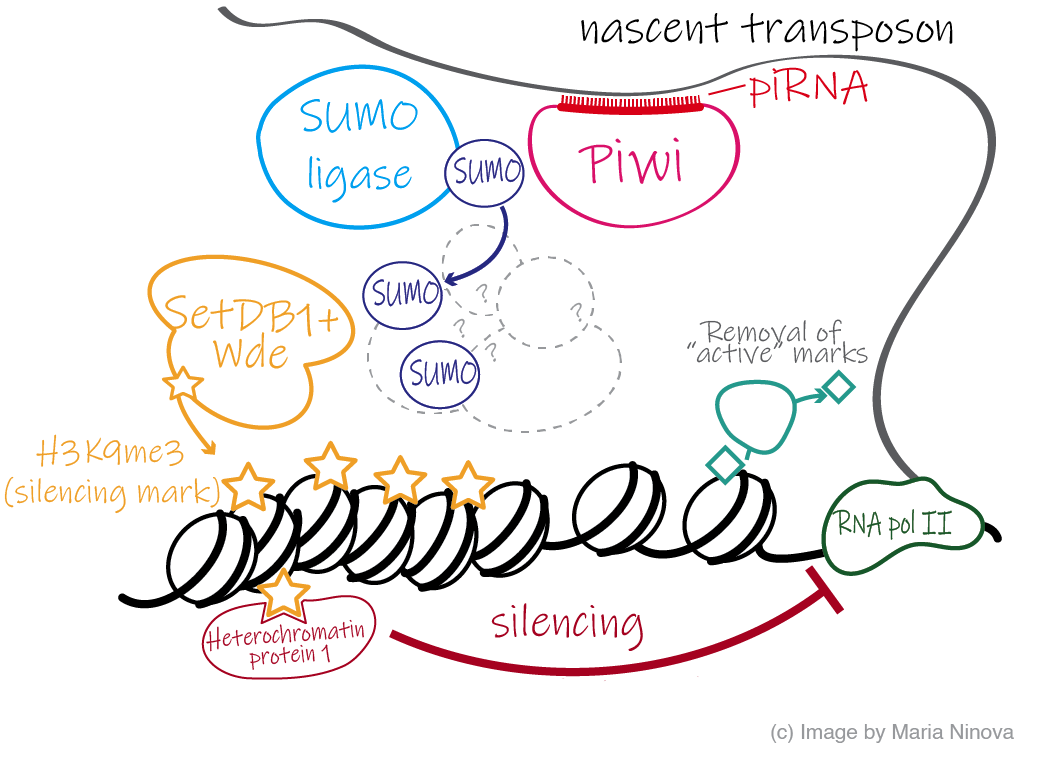 piRNAs are a class of short non-coding RNAs produced in the germline and some somatic tissues of animals that associate with Argonaute proteins from the Piwi clade, and function to recognize and silence transposons that can otherwise damage DNA. Defects in the piRNA pathways lead to transposon activation, defects in gametogenesis, and sterility. Remarkably, specific piRNA-loaded Piwi-s have the ability to enter the nucleus and induce transcriptional silencing of transposons by recruiting repressive chromatin-modifying factors - a great example of non-coding RNAs triggering epigenetic modifications in animal systems. Recent studies identified several new factors essential for Piwi function. For example, it recently emerged that
the SUMO (Small Ubiquitin-Like Modifier) pathway is required for transposon silencing and repessive chromatin formation at piRNA targets. However, the precise mechanism by which piRNA/Piwi complexes recruit the silencing effectors is not well understood. Our lab aims to use a combination of molecular biology, genomics, and proteomics strategies, to dissect the process of piRNA-mediated silencing.
piRNAs are a class of short non-coding RNAs produced in the germline and some somatic tissues of animals that associate with Argonaute proteins from the Piwi clade, and function to recognize and silence transposons that can otherwise damage DNA. Defects in the piRNA pathways lead to transposon activation, defects in gametogenesis, and sterility. Remarkably, specific piRNA-loaded Piwi-s have the ability to enter the nucleus and induce transcriptional silencing of transposons by recruiting repressive chromatin-modifying factors - a great example of non-coding RNAs triggering epigenetic modifications in animal systems. Recent studies identified several new factors essential for Piwi function. For example, it recently emerged that
the SUMO (Small Ubiquitin-Like Modifier) pathway is required for transposon silencing and repessive chromatin formation at piRNA targets. However, the precise mechanism by which piRNA/Piwi complexes recruit the silencing effectors is not well understood. Our lab aims to use a combination of molecular biology, genomics, and proteomics strategies, to dissect the process of piRNA-mediated silencing.
Regulation of gene expression by H3K9me3
Heterochromatin was historically identified as the dark staining portion of interphase chromatin which remains condensed and is typically (but not always) transcriptionally silenced. Heterochromatin is essnetial for many aspects of cell function and development, such as dosage compensation, lineage-specific gene silencing during differentiation, maintaining the integrity of the repetitive chromosome ends, and repression of transposons. Histone 3 lysine 9 trimethylation (H3K9me3) is a highly conserved hallmark of silencing and heterochromatin that occupies a substantial portion of the eukaryotic genomes and is implicated in the compaction of repeat-rich sequences near centromeres and telomeres, and transposon repreression. In addition, this mark is found on dozens of host genes and regulatory regions outside constitutive heterochromatin, and appears to play a role in
gene regulation and cell specification. Our lab is interested in gaining a deeper understanding of the biological role of H3K9me3 in different cellular and genomic contexts.
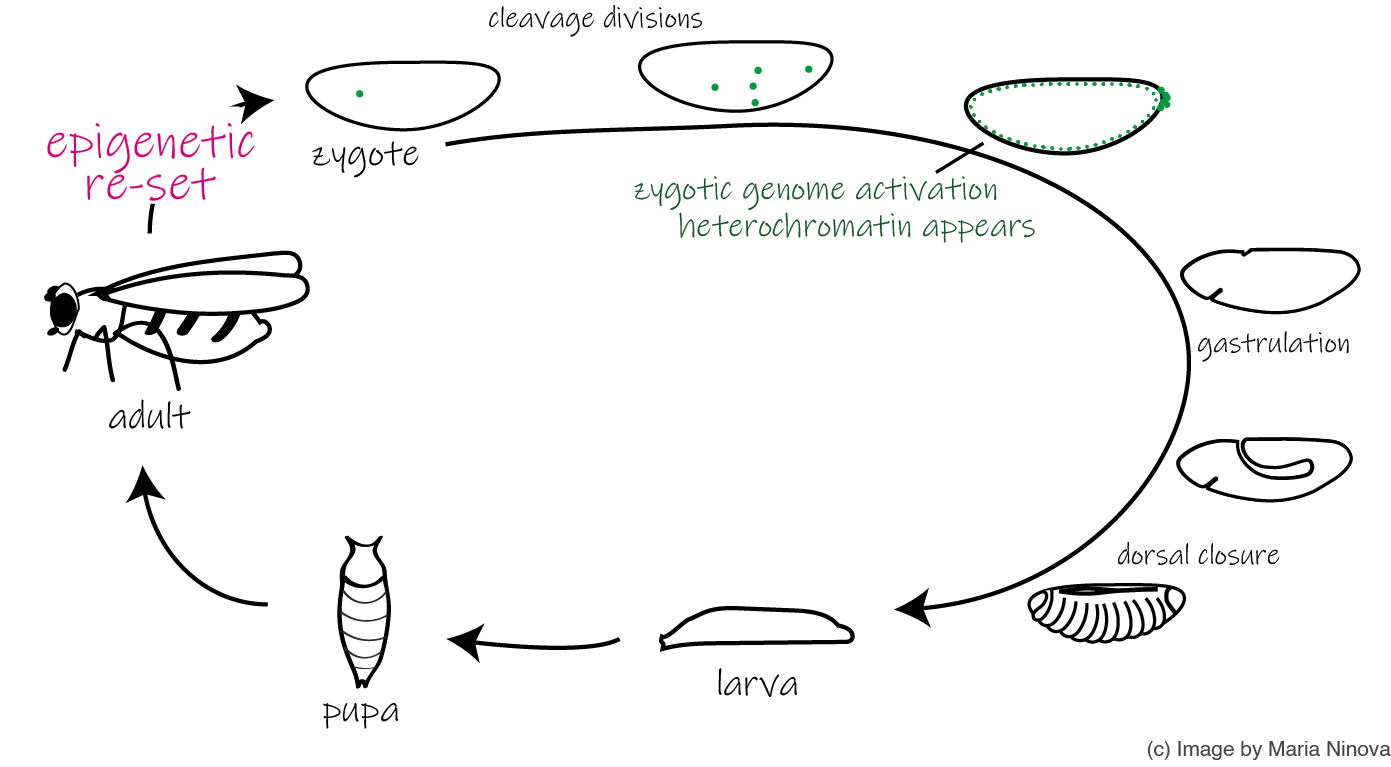
Developmental regulation of heterochromatin and transposabe elements
The vast majority of histone and DNA modifications are erased during gametogenesis and fertilization, and need to be re-established de novo in a cell-specific manner during the development of the new organism. We are interested in the factors and molecular mechanisms that orchestrate the initiation and maintenance of heterochromatin at host genes and transposons from the zygote throughout embryogenesis, as well as the functional implications of transposon activity, using the fruit fly embryo system as a paradigm.